We are looking for motivated postdocs and students to join the team!
Email Dr. Ma directly if you are interested. [read more…]

2026.02 Together with Dr. Jun Wang’s group at Stony Brook, we are awarded an R21 by NIA!
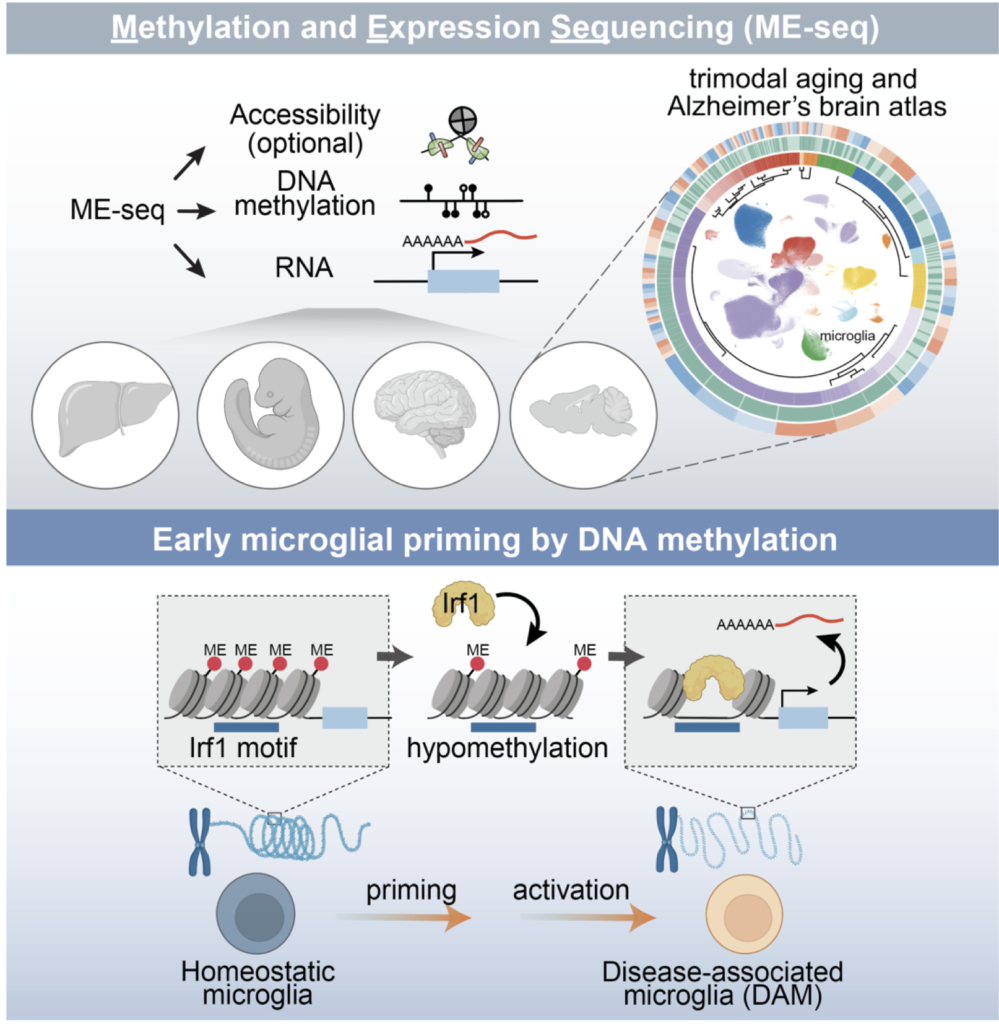
2026.02 Congratulations to Bohan and team for putting together this exciting work! We engineered ME-seq for ultra-high throughput single-cell DNA methylation & RNA co-mapping and identified methylation-mediated priming in aging and Alzheimer’s brain! [read more…]
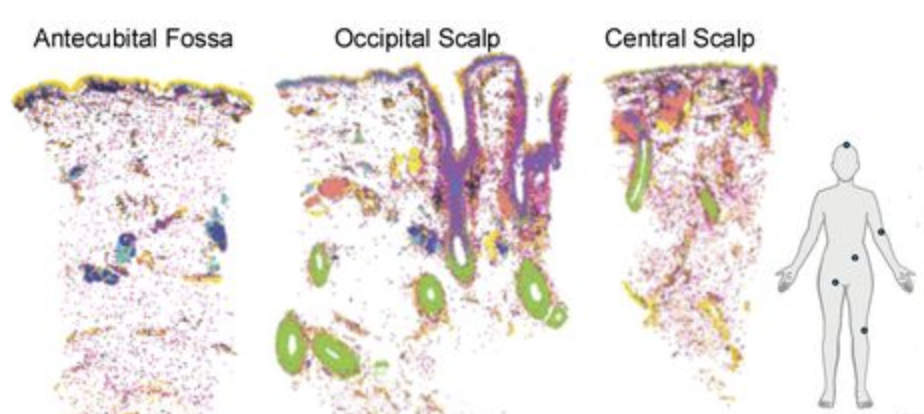
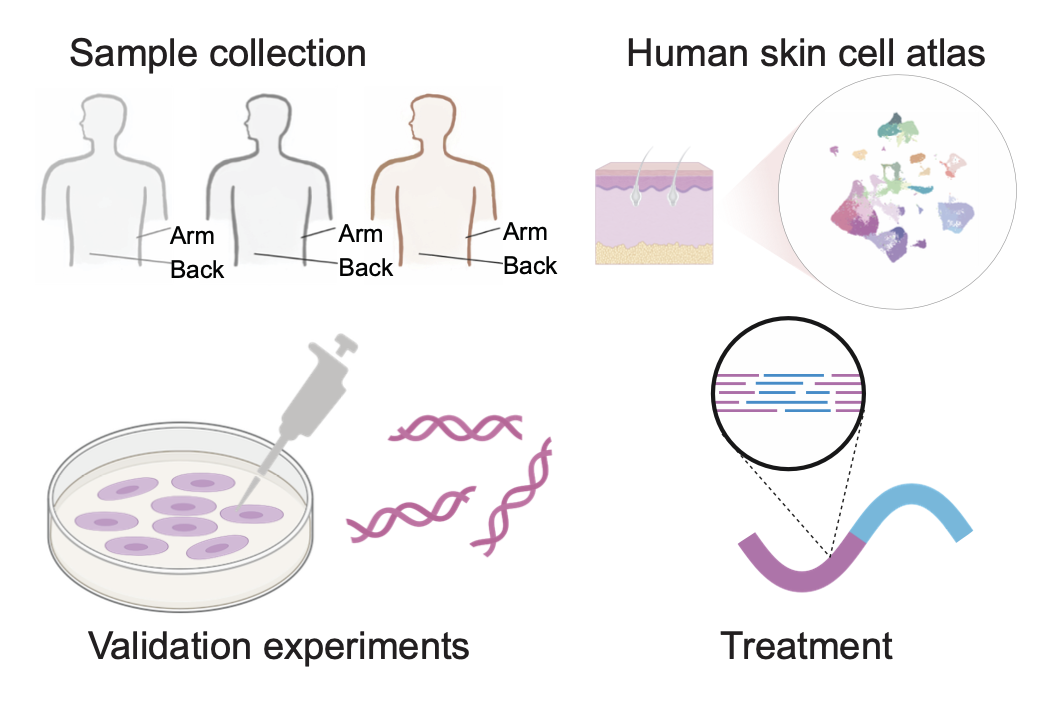
2025.12 The spatial human skin atlas paper mapping multiple anatomic sites led by Dr. Andrew Ji’s group is now accepted in Nature Genetics! [read more…]

2025.09 Welcome Dawei Tang, who is joining us as a master student to tackle brain functions!
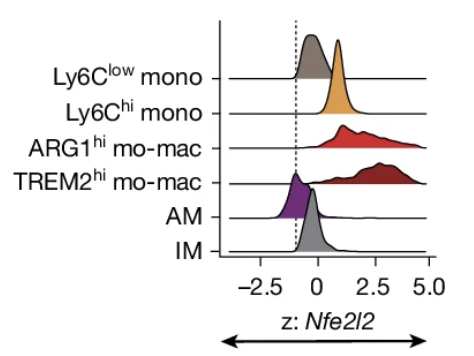
2025.09 Our collaborative work with Dr. Miriam Merad’s group is now published in Nature! We show that lung tumours prime accessibility for Nfe2l2 (NRF2) in bone marrow myeloid progenitors, enhancing myelopoiesis while dampening interferon response and promoting immunosuppression.

2025.08 We are excited to partner with Dr. Elvin Wagenblast’s group on an R21-funded project to address Down syndrome.

2025.07 We just celebrated a major milestone—our first PhD student, Dr. Federico Di Tullio, has officially graduated from the lab! Congratulations, Fred! Wishing you all the best as you start your next chapter at UCSD.

2025.07 Welcome back Ashley Chan as a returning summer intern!

2025.06 Huge congrats to Brian Soong on his NCI/NIH F30 award. Incredibly proud of you!
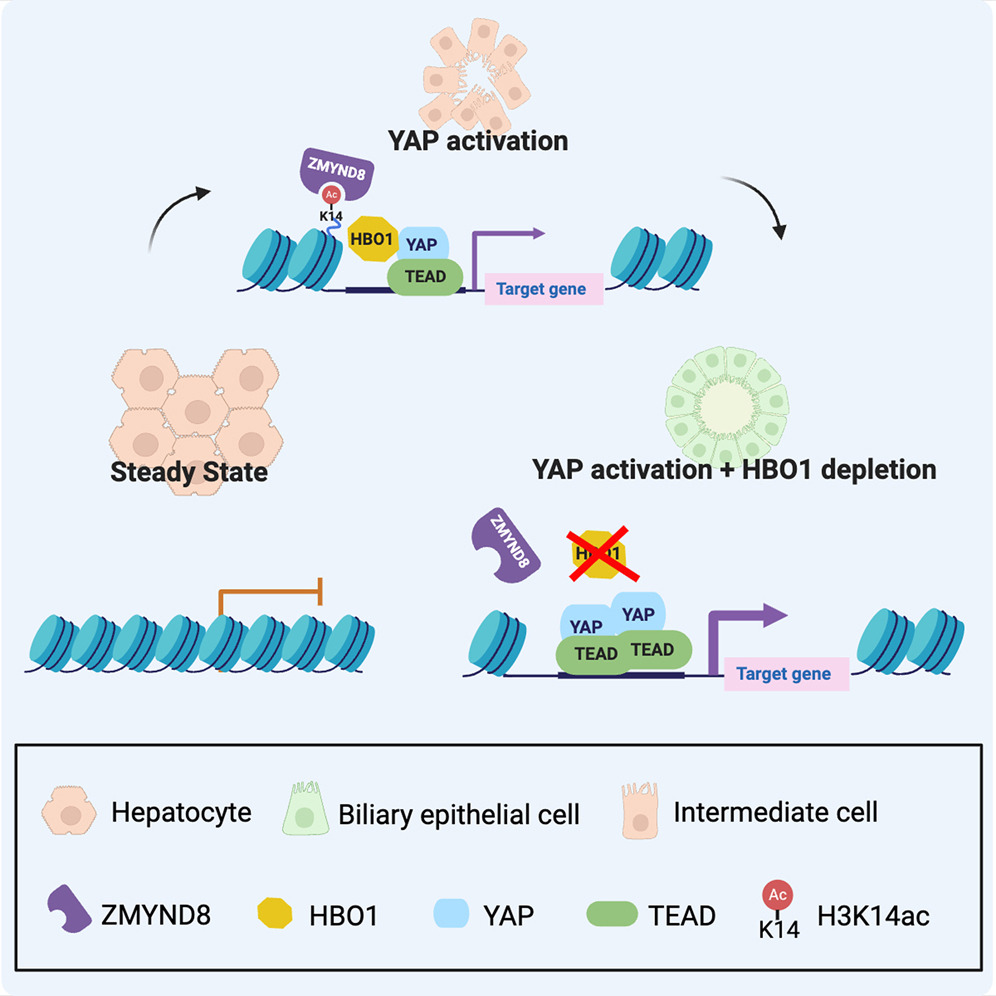
2025.05 Our work with Wei-Chien Yuan, Fernando Camargo and my postdoc mentor Jason Buenrostro is published in Cell Stem Cell. We leveraged in vivo CRISPR screen and single-cell ATAC to identify HBO1 as a critical barrier to hepatocyte reprogramming. [read more…]

2025.05 Our previous intern Jiayi Li is accepted as a master student in computational biology at Harvard! Congratulations!

2025.03 Our lab is now funded by NIH/NCI IMAT program to study AML!

2024.12 Partnered with Herbert Wu, we are honored to be selected as 2025 FBI scholars!
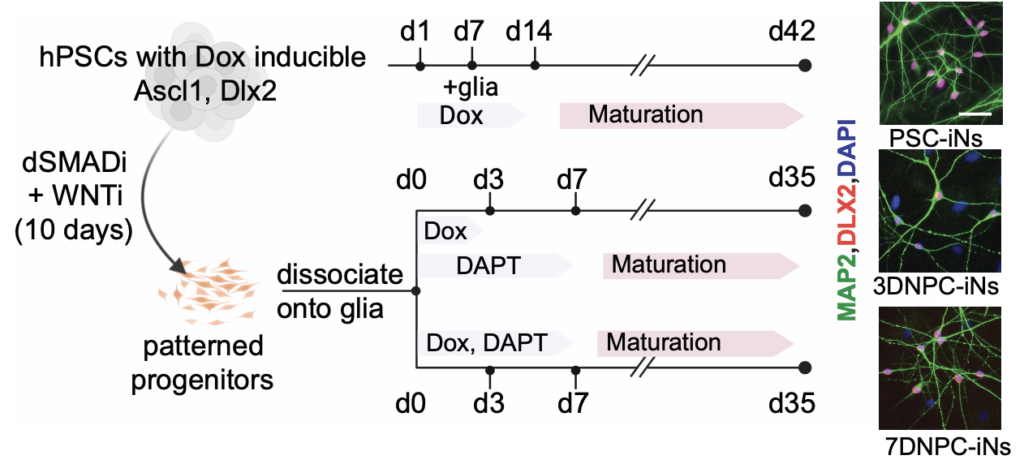
2024.11 With close colleborator Dr. Nan Yang’s team, we designed a new strategy to induce GABAergic neurons for better disease modeling and leveraged single-cell assays to probe the heterogeneity. [read more…]

2024.11 Our footprinting paper is accepted in Nature. We use machine learning to correct Tn5 insertion bias, enabling accurate prediction of TF binding in large-scale scATAC datasets.[read more…]
2024.11 Together with George Church’s Lab, we found a new TF ATF3 for rejuvenating human skin [read more…]

2024.11 Teamed with Dr. Elvin Wagenblast, we are awarded by the Henry & Marilyn Taub Foundation for a pilot award.

2024.06 Congratulations to our previous intern, Hyeonseo Lee, got accepted into WUSTL as a pre-med student! Super proud of you!

2024.06 We are awarded by Melanoma Research Alliance, together with Anne Bowcock, Elena Ezhkova and Jose Silva’s groups
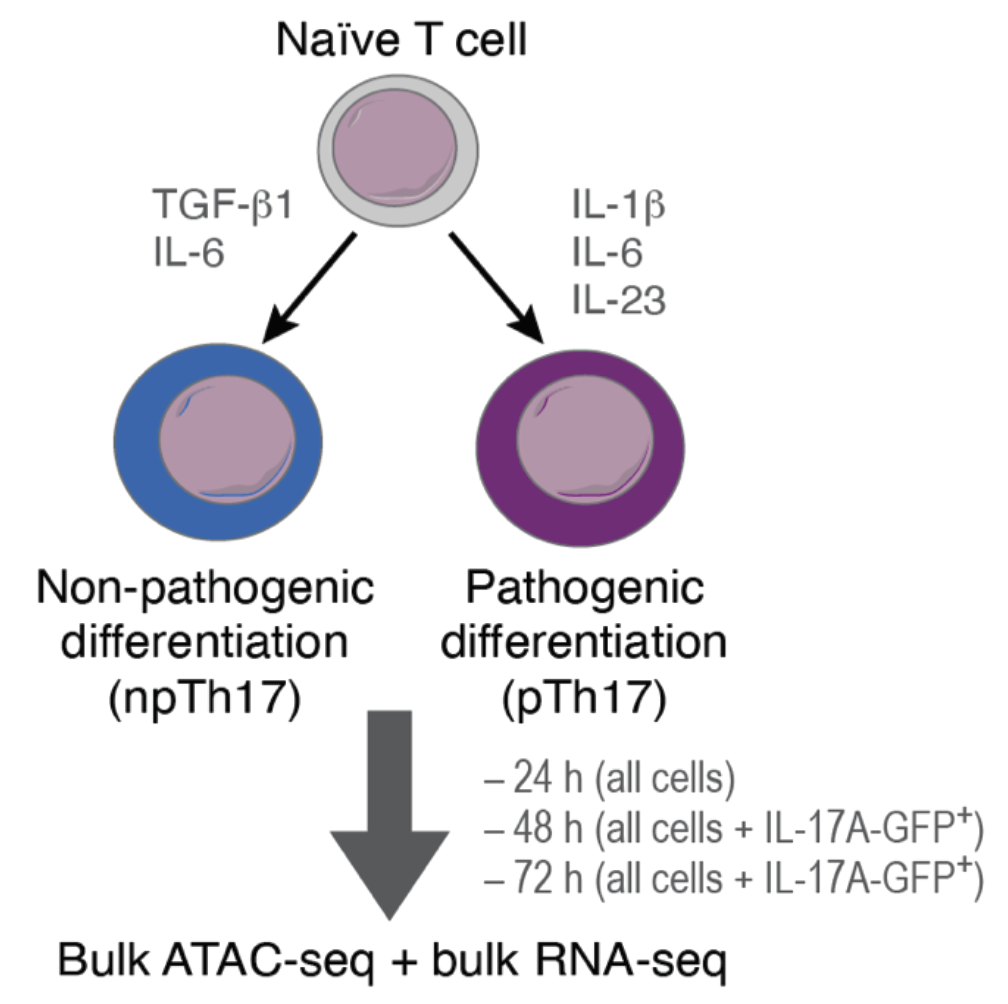
2024.05 Our collaborative paper with Aviv Regev and Vijay K. Kuchroo on Th17-based autoimmunity is now accepted in Nature Immunology! [read more…]

2024.01 Welcome visiting PhD student Liying Wang join our lab!

2024.01 Welcome PhD student Lillian Chang to rotate in our lab.

2023.08 Welcome PhD student Arushi Samal to rotate in our lab.

2023.08 Welcome PhD student Nazifa Salsabeel to rotate in our lab.

2023.08 – Our collaborative paper with Duygu Ucar lab and Steven Z. Josefowicz lab is now accepted in Cell. Transcriptomic and epigenomic analysis of blood reveal sustained changes in hematopoiesis and innate immunity after COVID-19. [read more…]

2023.07 Brian Soong joins our lab as a MD/PhD student! He will be co-mentored by Miriam Merad to investigate the epigenetic clue in the immune environment.

2023.07 Hyeonseo Lee joins our lab as a summer intern! Welcome!
2023.07 – Sai is now a member of the Tisch Cancer Institute (TCI) with the goal of advancing clinical breakthroughs that help prevent and eradicate cancer.

2023.07 – Welcome PhD student Federico Di Tullio joining our lab! He will work on liver injury model and be co-mentored with Dr. Tianliang Sun.

023.06 – Our collaborative paper with Dr. Chang Lu at Virginia Tech is now accepted in Nature Communications. We demonstrate a droplet-based microfluidic technology, Drop-BS, to construct single-cell bisulfite sequencing libraries for DNA methylome profiling. Check out the paper! [read more…]

2023.05 – Welcome PhD student, Zhicong (Leon) Liao, officially joined our lab!

2023.04 – Our collaborative work with Cigall Kodach’s group is now published in Molecular Cell! We leveraged CRISPR-Cas9 knockout screens to target mSWI/SNF subunits individually and in select combinations, followed by single-cell RNA-seq and multi-mic assay (perturb-SHARE-seq). These single-cell subunit perturbation signatures mapped across bulk primary human tumor expression profiles both mirror and predict cBAF loss-of-function status in cancer. [read more…]

2023.03 – Our are preprint is now on bioRxiv! We present a computational tool, PRINT, to predict the chromatin binding proteins (TF & histone) from large-scale scATAC data . [read more…]

2023.03 – Our collaborative paper with Dr. Rong Fan at Yale University is now published in Nature. We present two technologies for spatially resolved, genome-wide, joint profiling of the epigenome and transcriptome by co-sequencing chromatin accessibility/histone modification and gene expression on the same tissue section at near-single-cell resolution. [read more…]

2023.03 – Welcome PhD student, Zhicong (Leon) Liao, rotating in our lab!

2023.02 – Dr. Feifei Yuan joined our lab as a new postdoctoral fellow. Welcome!

2023.01 – Our collaboration with Feng Zhang’s group at Broad Institute is now published in Cell. We massive screened every single TF in hESC differentiation by combining SHARE-seq with genetic perturbation and surprisingly found 27% of TF genes could function as master regulators. [read more…]

2022.11 – Dr. Ma is selected as an Affiliate Member of the New York Genome Center (NYGC).

2022.10 – Welcome Nima Assad joining the lab as the first MD/PhD student!

2022.09 – Welcome our first postdoctoral fellow Dr. Bohan Zhu to the lab!

2022.08 – Ma Lab officially opens in the Department of Genetics and Genomic Sciences at Icahn School of Medicine at Mount Sinai! Join us and work on cool science in the heart of Manhattan! [read more…] & [read more…]

2022.08 – Our collaborative work is published in Nature Neuroscience. We aim to investigate the regulatory strategies in both early post-mitotic and later stages of neurons from mouse and marmoset neocortex. [read more…]

2022.06 – We seek to incorporate spatial information with chromatin accessibility for mapping various tissues! Now accepted in Nature. [read more…]
- A collaboration with Dr. Rong Fan’s group at Yale

2022.06 – FigR-a very well optimized computational workflow to pair scATAC to scRNA data, and infer gene regulation network for stimulated PBMCs. Now accepted in Cell Genomics. [read more…]
- Lead by Dr. Vinay Kartha and Dr. Fabiana Duarte
- PBMC stimulation data exploration

2022.02 – We use a microfluidic approach, MOWChIP-seq, to investigate the cell-type-specific impact of BRCA1 mutation on human breast tissue. [read more…]

2021.09 – Our work on investigating the loss of Smarca4 that results in highly advanced dedifferentiated tumors is now in Cancer Discovery! [read more…]

2021.06 – Gata2b-deficient zebrafish recapitulate human GATA2 deficiency syndrome–associated hematopoietic phenotypes. [read more…]
- A collaboration with Dr. Len Zon’s group at HMS

2021.06 – Our optimized sci-ATAC protocol is now available in STAR protocols [read more…]
- A collaboration with Isabella Del Priore and Lindsay LaFave in Tyler Jacks’ Lab at MIT.

2021.04 – Our work to understand chronic stress that leads to hair loss is in Nature now [read more…]!
- A collaboration with Dr. Ya-Chieh Hsu’s group at Harvard, led by Dr. Sekyu Choi.

2021.02 – An interesting story about superenhancer-induced ELF3 activation in trophoblasts is now in PNAS [read more…].
- A collaboration with Dr. Jack L. Strominger’s group at Harvard, led by Dr. Qin Li.






