Developing cutting-edge single-cell sequencing approaches to probe gene regulation mechanisms and cell fate transitions in development and diseases
Technology development
We are keen on developing sequencing technologies to probe epigenenetic landscapes in tiny amounts of samples, single-cells and in situ, which provides a means to better understand the gene regulation mechanisms in complex tissues. We have developed SurfaceChIP and MID-RRBS for ultra-sensitive genome-wide histone modification and DNA methylation profiling from hundreds of cells. We also developed simultaneous high-throughput ATAC and RNA expression with sequencing (SHARE-seq), a powerful powerful approach for co-profiling chromatin and RNA from the same single cells. We are actively developing tools to further understand the interaction between different modalities (multi-‘omics’) and its impact on cell behavior.
8 .
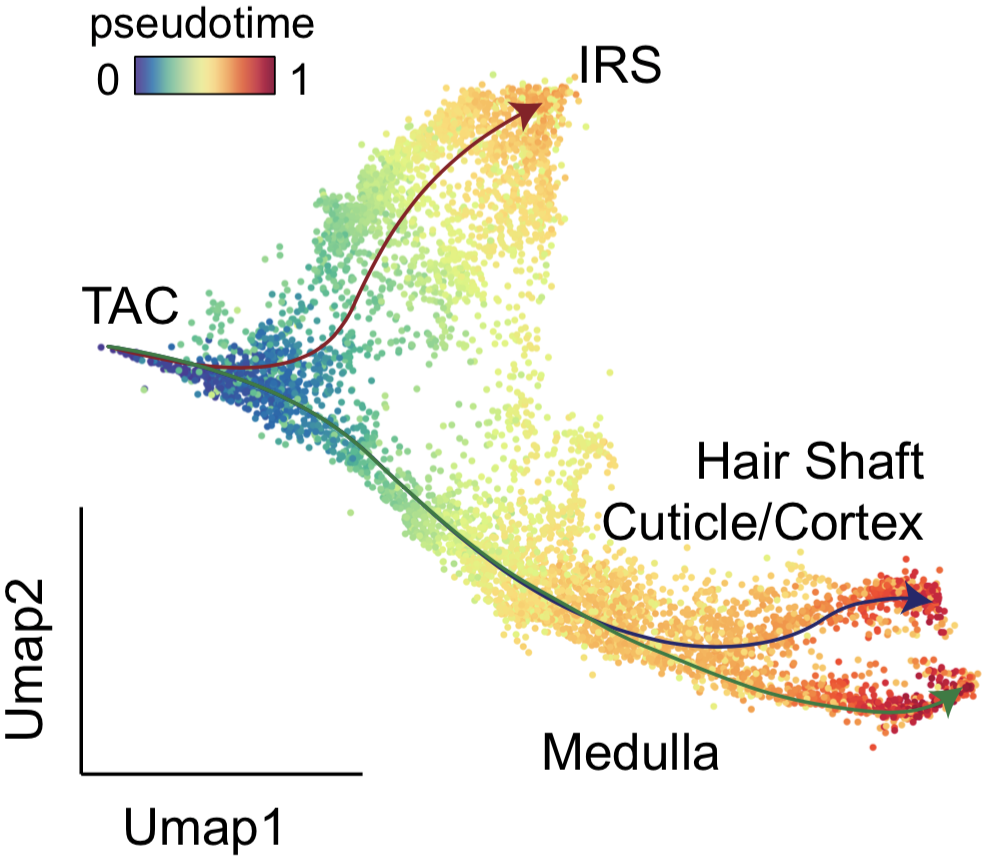
Gene regulation inference in cell differentiation
One of the most fundamental questions in biology is to understand how the cells spontaneously grow and proliferate, and how the cells respond to external stimulation. For example, when experiencing chronic stress or acute stress, the diverse skin populations respond quite differently which leads to hair loss and hair greying. Leveraging sequencing technologies and other approaches, we demonstrated the molecular driver underlying the stress response. We also developed a computational approach, Chromatin potential, to infer the lineage-fate choice of skin stem cells during cell fate transition. I will continuously investigate the mechanism of cell fate shift by building new computational tools and exploring the cell behavior under other stimulations.
.
.
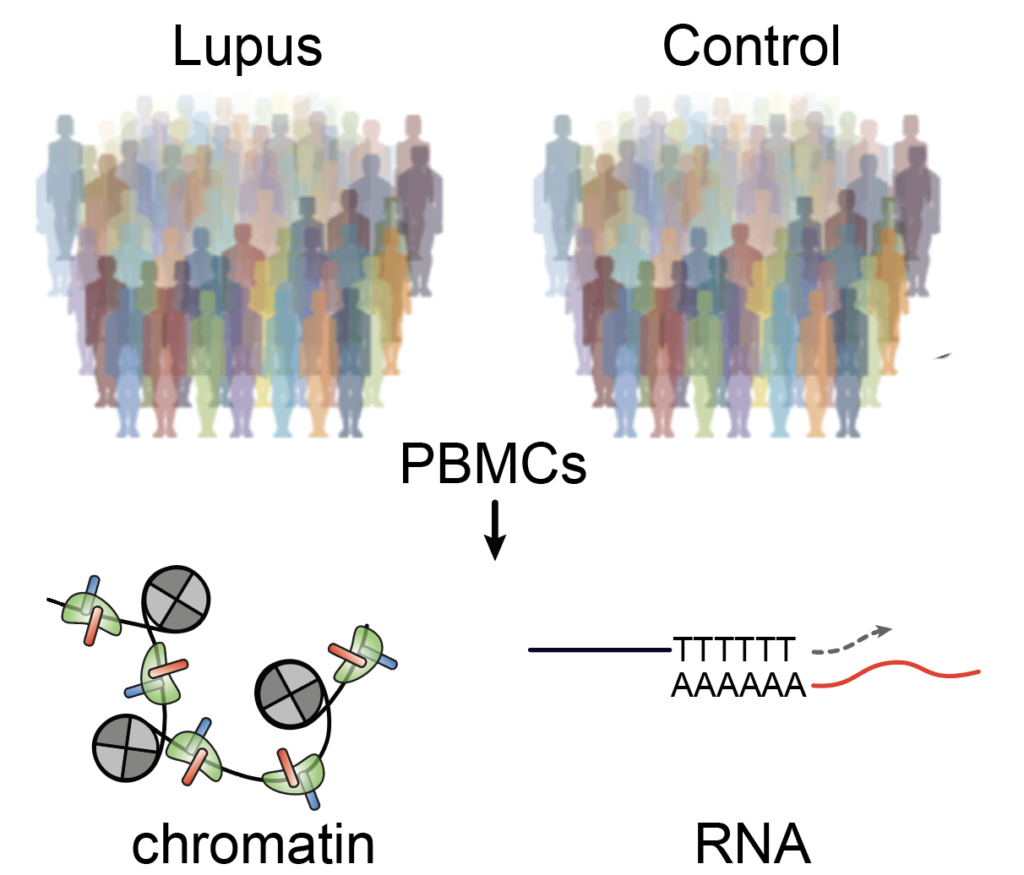
Genetic clues in autoimmune diseases and cancers
Autoimmune diseases, including lupus, are caused by the immune system mistakenly attacks the body. The vast majority of disease-associated loci are located in non-coding regions of the genome. Genetic analysis of variations in chromatin state is a powerful approach for identifying SNPs that directly affect cis-regulatory activity. Leveraging single-cell chromatin measurements lupus patients, we built a relationship between genetic variants, chromatin accessibility, and transcriptional regulation. Separately, we also demonstrated that the epigenomic programs, specifically, lineage TFs bias the cell fate outcomes in lung tumors. We seek to use these findings to further understand other immune diseases and cancers.

